Code
library(bggjphd)
library(tidyverse)
library(GGally)
library(cowplot)
library(arrow)
library(here)
library(future)
library(progressr)
theme_set(theme_bggj())In our case we want \(\mu\) to vary with time. If we write \(y_{it}\) for the observed hourly maximum at station \(i\) during year \(t\) we will then have
where
and
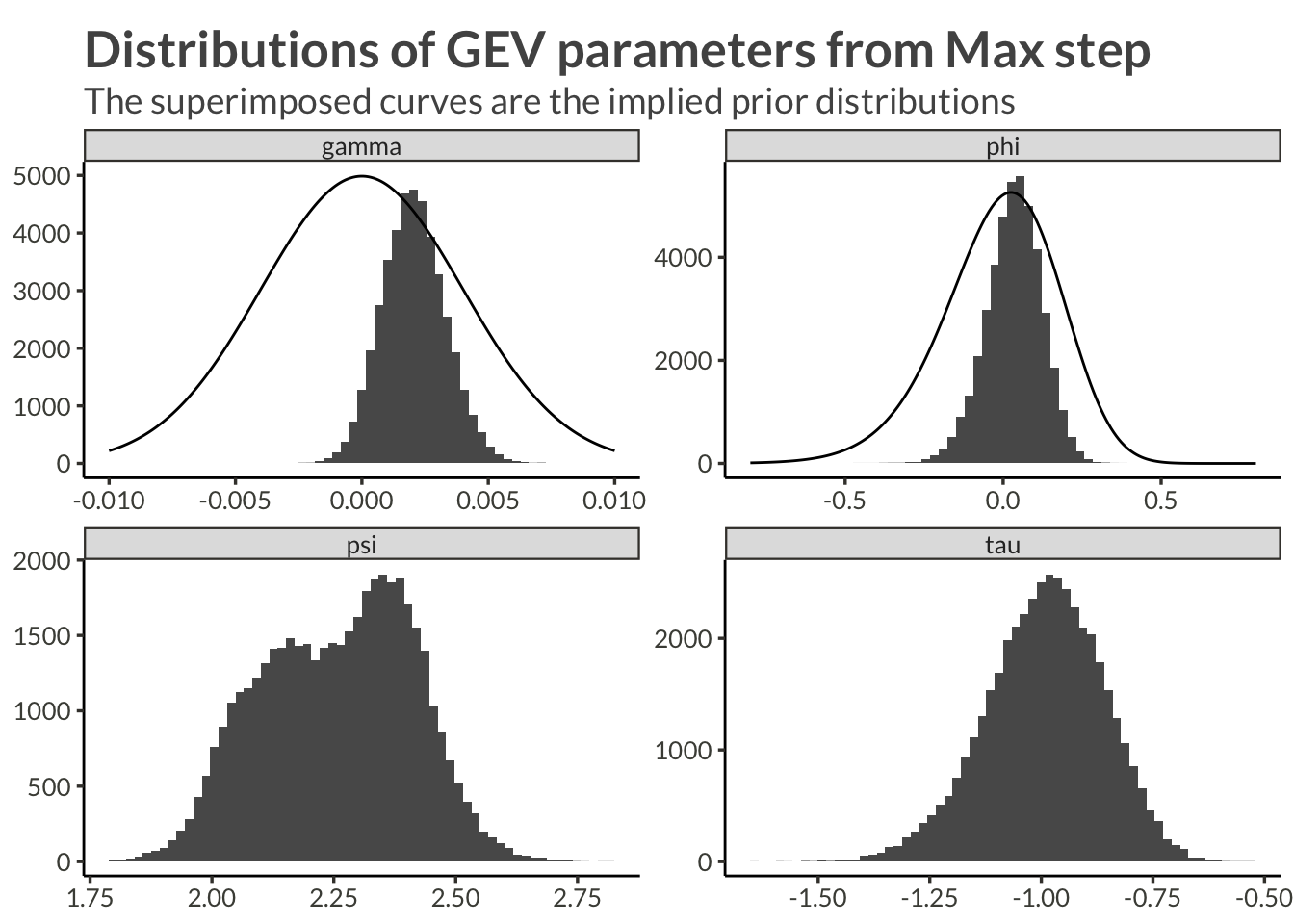
Having performed the Max step and saved the ML estimates we can easily load them by fetching the object station_estimates. Here we plot the distribution of transformed estimates.
where the link functions for the shape and trend parameters are defined according to Johannesson et al. (2021)
d_prior <- crossing(
name = c("phi", "gamma"),
x = seq(-1, 1, len = 200)
) |>
mutate(
x = ifelse(name == "phi", x * 0.8, x * 0.01),
y = ifelse(name == "phi", prior_shape(x) |> exp(), prior_trend(x) |> exp()),
y = ifelse(name == "phi", y * 6000, y * 0.05)
)
d |>
ggplot(aes(value)) +
geom_histogram(bins = 60) +
geom_line(
data = d_prior,
aes(x = x, y = y * 1e3),
inherit.aes = F
) +
facet_wrap("name", scales = "free") +
labs(
x = NULL,
y = NULL,
title = "Distributions of GEV parameters from Max step",
subtitle = "The superimposed curves are the implied prior distributions"
)
d_prior <- crossing(
name = c("phi", "gamma"),
x = seq(-1, 1, len = 200)
) |>
mutate(
x = ifelse(name == "phi", x * 0.8, x * 0.01),
y = ifelse(name == "phi", prior_shape(x) |> exp(), prior_trend(x) |> exp()),
y = ifelse(name == "phi", y * 2000000, y * 20),
x = ifelse(name == "phi", link_shape_inverse(x), link_trend_inverse(x)),
name = ifelse(name == "phi", "xi", "delta")
)
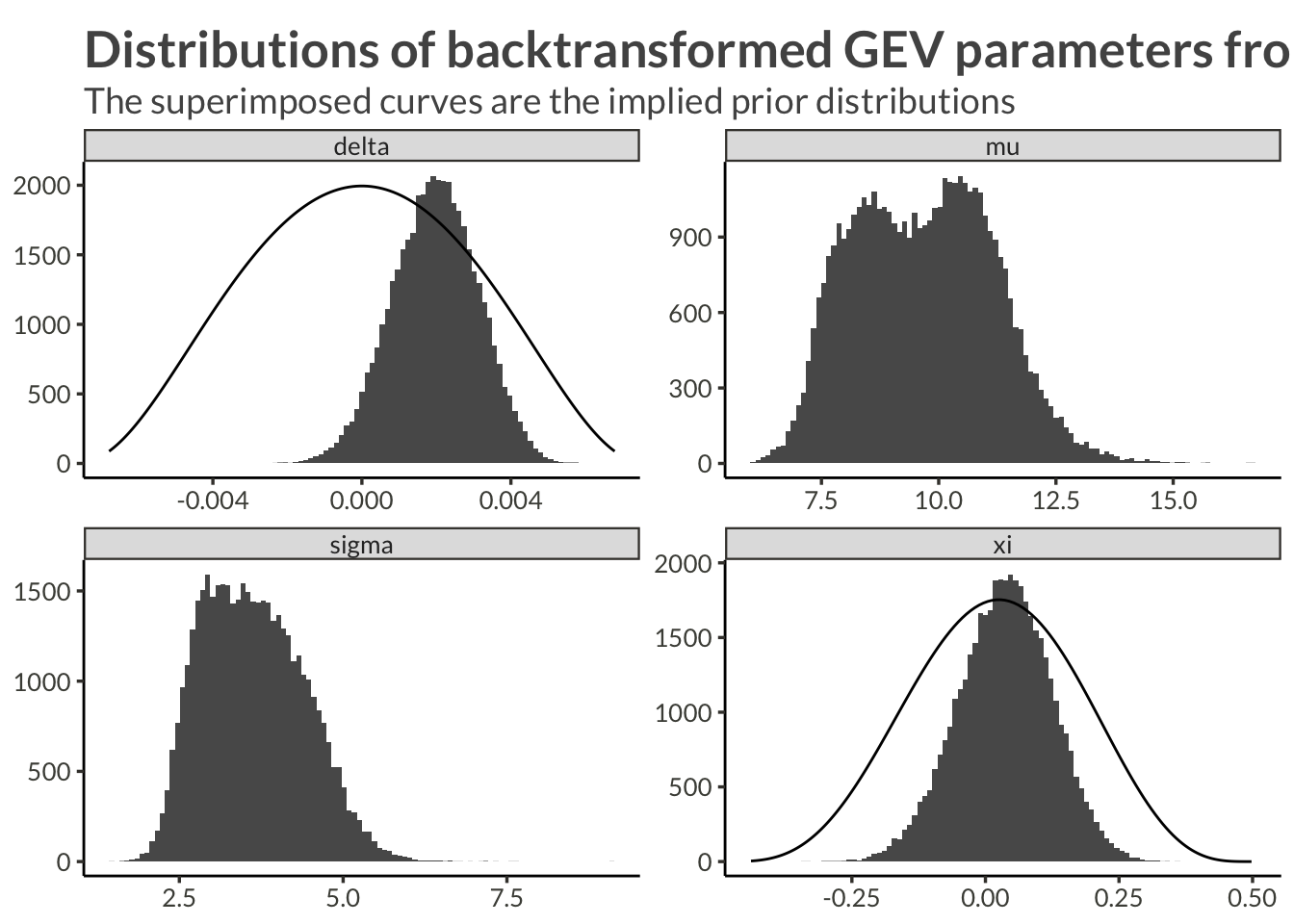
d |>
pivot_wider() |>
mutate(mu = exp(psi),
sigma = exp(tau + psi),
xi = link_shape_inverse(phi),
delta = link_trend_inverse(gamma)) |>
select(-psi, -tau, -phi, -gamma, -proj_x, -proj_y) |>
pivot_longer(c(-station)) |>
mutate(name = fct_relevel(name, "mu", "sigma", "xi", "delta")) |>
ggplot(aes(value)) +
geom_histogram(bins = 100) +
geom_line(
data = d_prior,
aes(x = x, y = y),
inherit.aes = F
) +
facet_wrap("name", scales = "free") +
labs(
x = NULL,
y = NULL,
title = "Distributions of backtransformed GEV parameters from Max step",
subtitle = "The superimposed curves are the implied prior distributions"
)
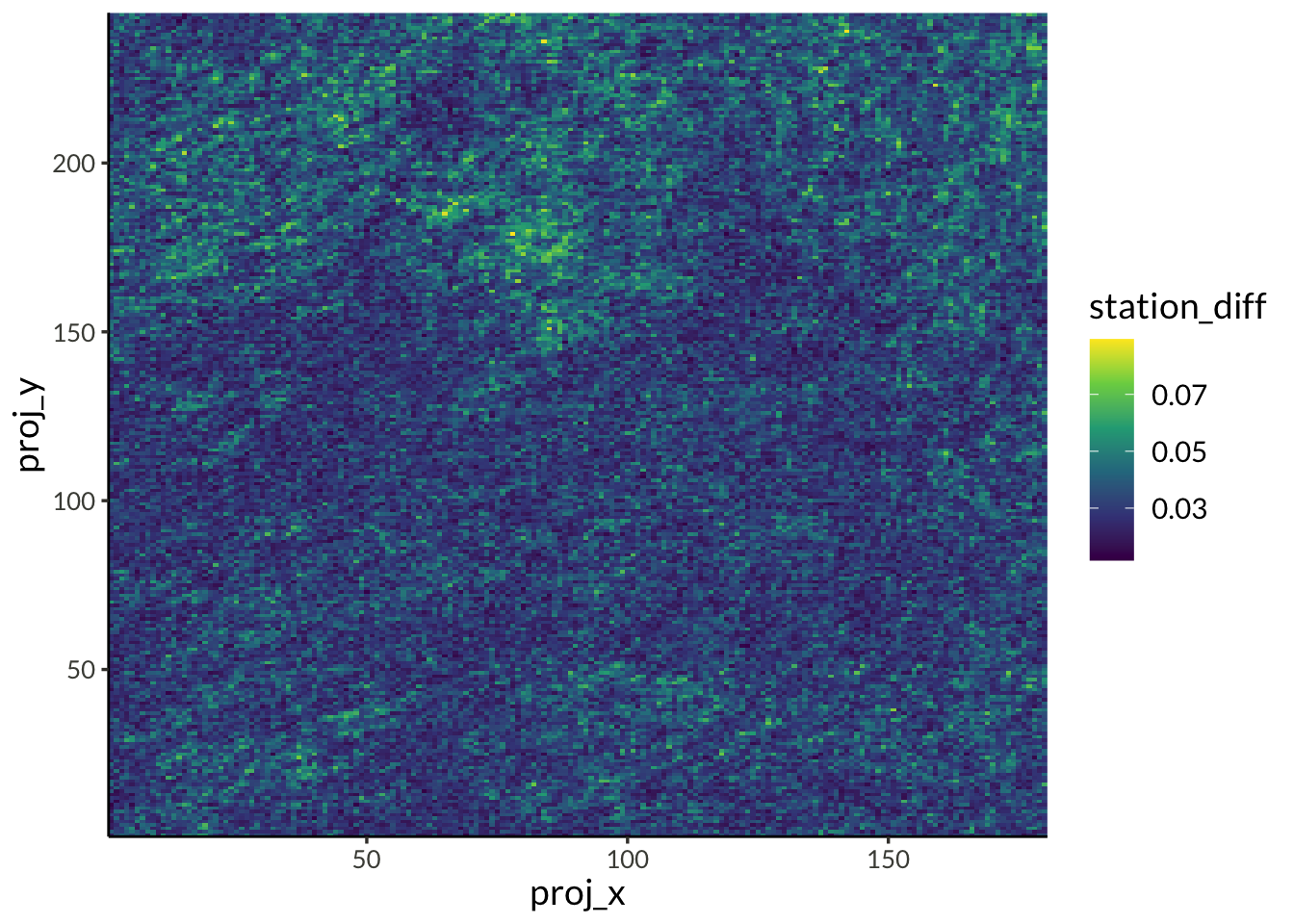
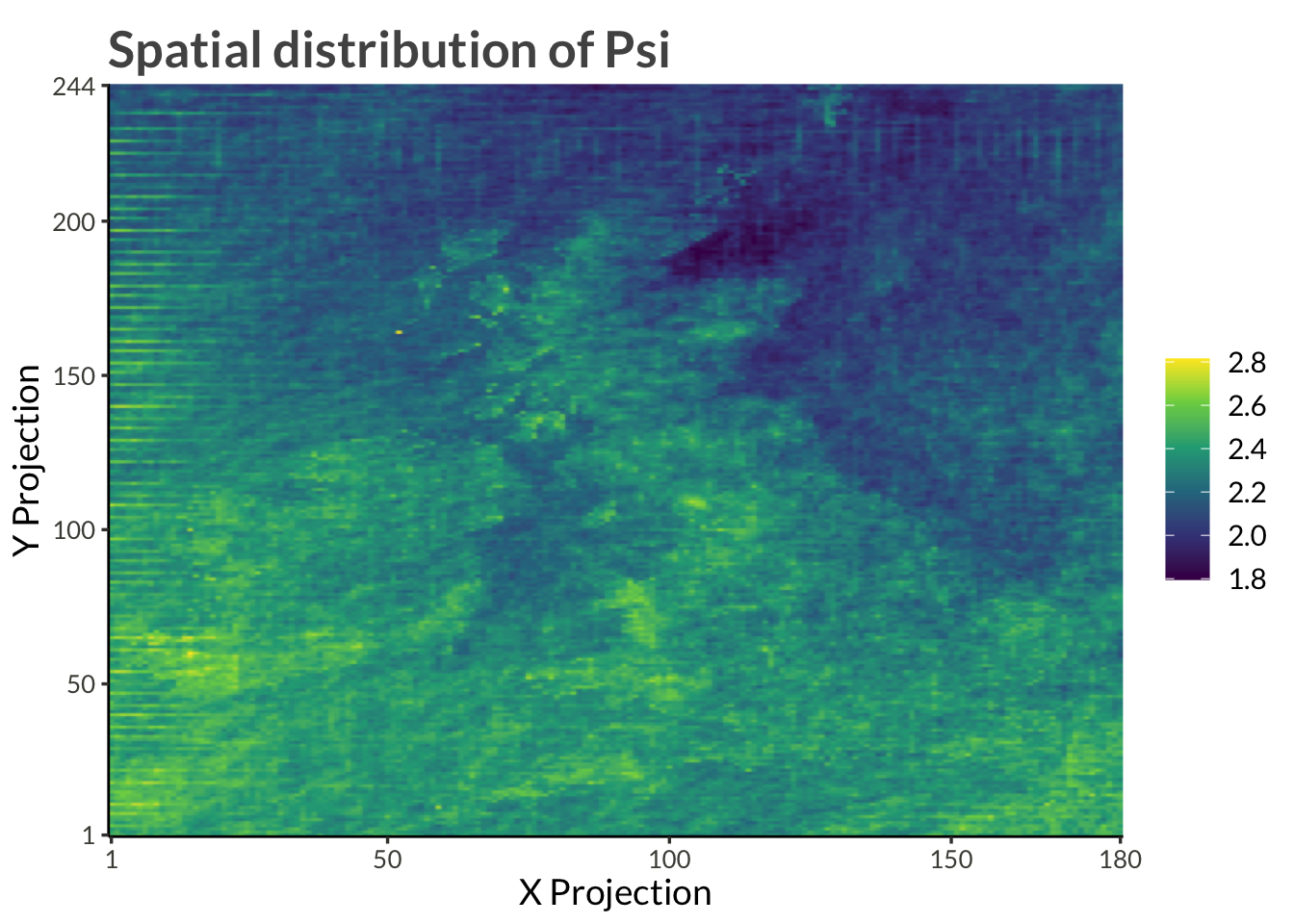
d |>
filter(name == "psi") |>
ggplot(aes(proj_x, proj_y, fill = value)) +
geom_raster(interpolate = TRUE) +
scale_x_continuous(
expand = expansion(),
breaks = c(range(d$proj_x), pretty(d$proj_x))
) +
scale_y_continuous(
expand = expansion(),
breaks = c(range(d$proj_y), pretty(d$proj_y))
) +
scale_fill_viridis_c() +
labs(
x = "X Projection",
y = "Y Projection",
fill = NULL,
title = "Spatial distribution of Psi"
)
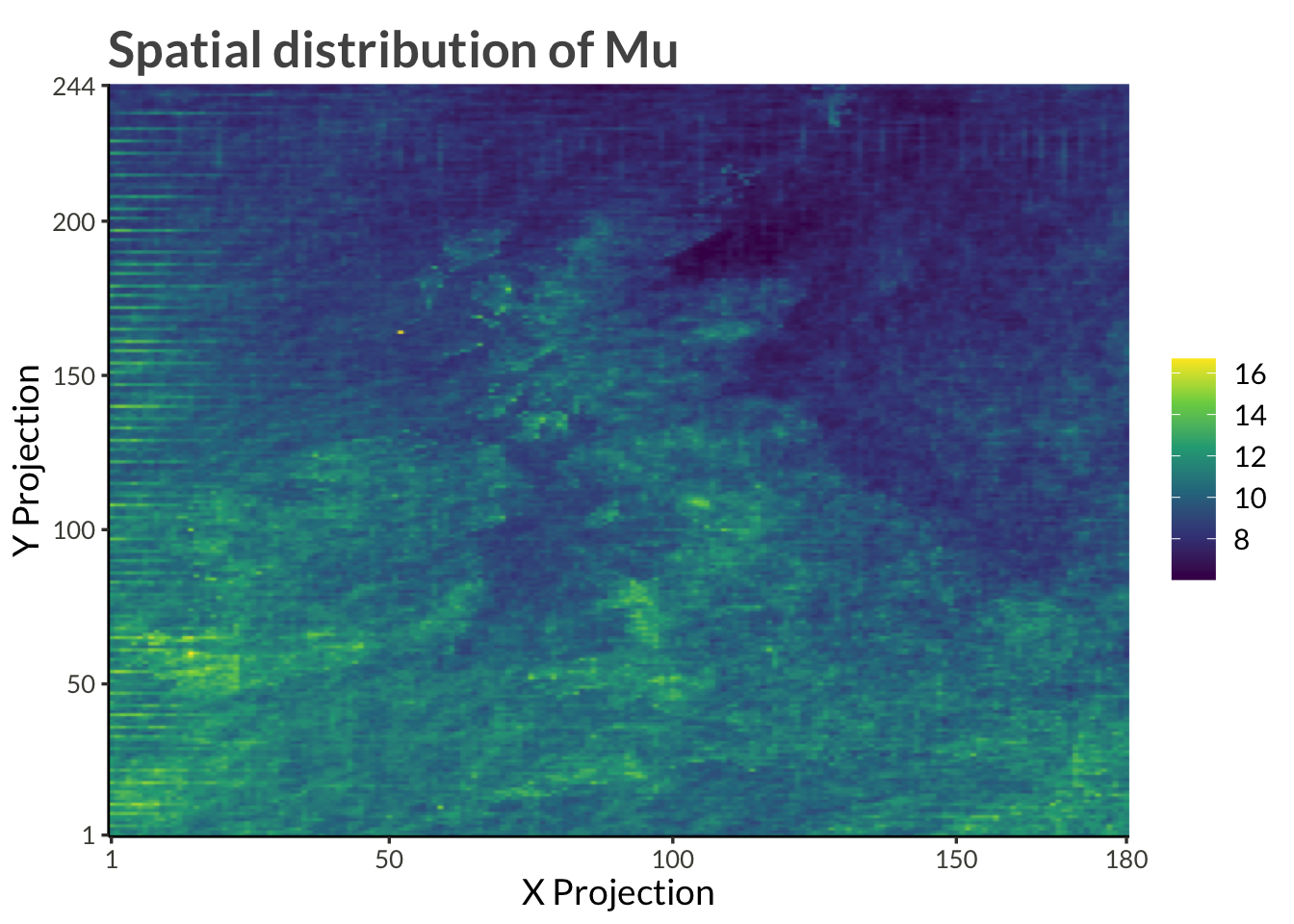
d |>
filter(name == "psi") |>
ggplot(aes(proj_x, proj_y, fill = exp(value))) +
geom_raster(interpolate = TRUE) +
scale_x_continuous(
expand = expansion(),
breaks = c(range(d$proj_x), pretty(d$proj_x))
) +
scale_y_continuous(
expand = expansion(),
breaks = c(range(d$proj_y), pretty(d$proj_y))
) +
scale_fill_viridis_c() +
labs(
x = "X Projection",
y = "Y Projection",
fill = NULL,
title = "Spatial distribution of Mu"
)
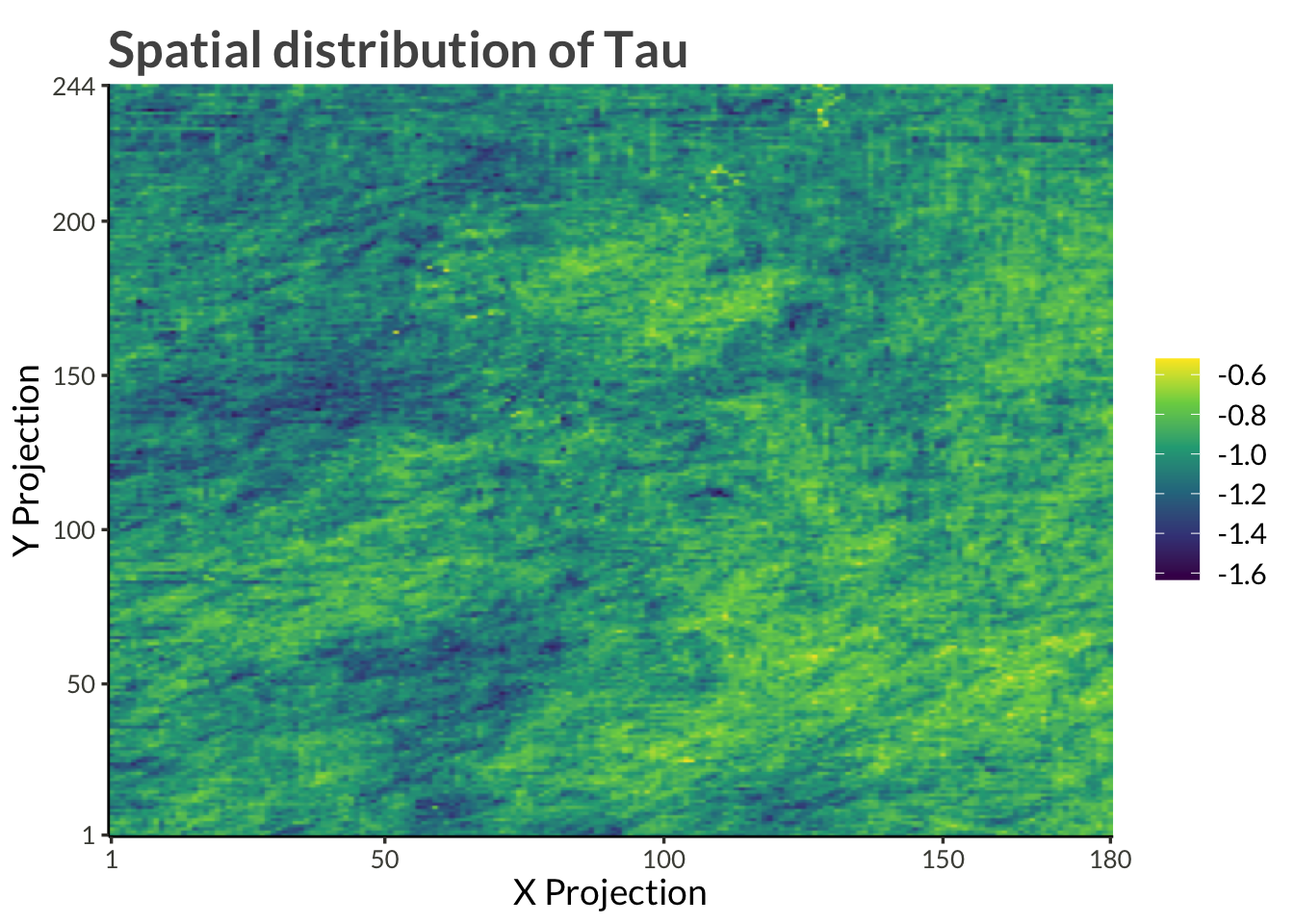
d |>
filter(name == "tau") |>
ggplot(aes(proj_x, proj_y, fill = value)) +
geom_raster(interpolate = TRUE) +
scale_x_continuous(
expand = expansion(),
breaks = c(range(d$proj_x), pretty(d$proj_x))
) +
scale_y_continuous(
expand = expansion(),
breaks = c(range(d$proj_y), pretty(d$proj_y))
) +
scale_fill_viridis_c() +
labs(
x = "X Projection",
y = "Y Projection",
fill = NULL,
title = "Spatial distribution of Tau"
)
d |>
pivot_wider() |>
mutate(sigma = exp(tau + psi)) |>
ggplot(aes(proj_x, proj_y, fill = sigma)) +
geom_raster(interpolate = TRUE) +
scale_x_continuous(
expand = expansion(),
breaks = c(range(d$proj_x), pretty(d$proj_x))
) +
scale_y_continuous(
expand = expansion(),
breaks = c(range(d$proj_y), pretty(d$proj_y))
) +
scale_fill_viridis_c() +
labs(
x = "X Projection",
y = "Y Projection",
fill = NULL,
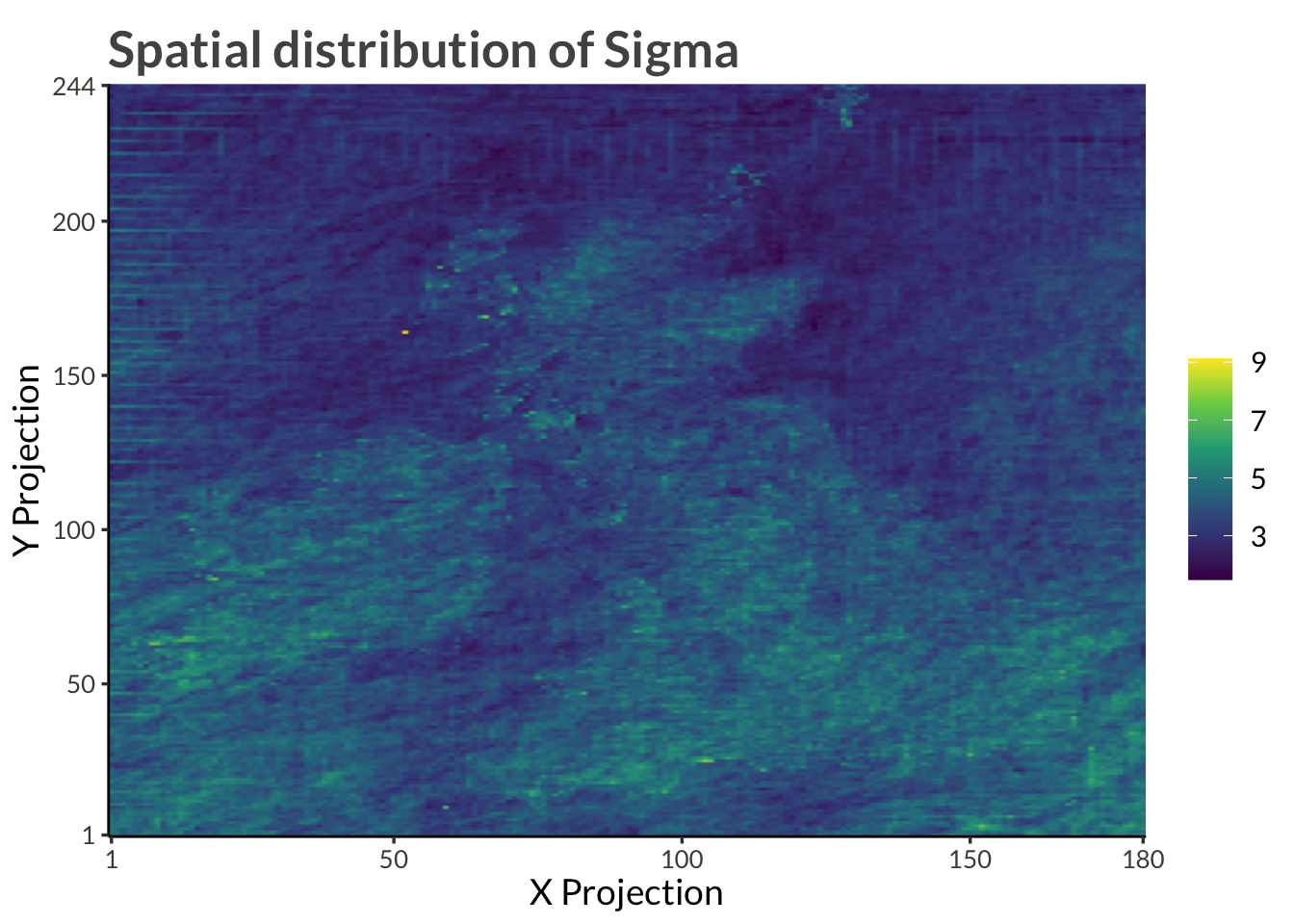
title = "Spatial distribution of Sigma"
)
d |>
filter(name == "phi") |>
ggplot(aes(proj_x, proj_y, fill = value)) +
geom_raster(interpolate = TRUE) +
scale_x_continuous(
expand = expansion(),
breaks = c(range(d$proj_x), pretty(d$proj_x))
) +
scale_y_continuous(
expand = expansion(),
breaks = c(range(d$proj_y), pretty(d$proj_y))
) +
scale_fill_viridis_c() +
labs(
x = "X Projection",
y = "Y Projection",
fill = NULL,
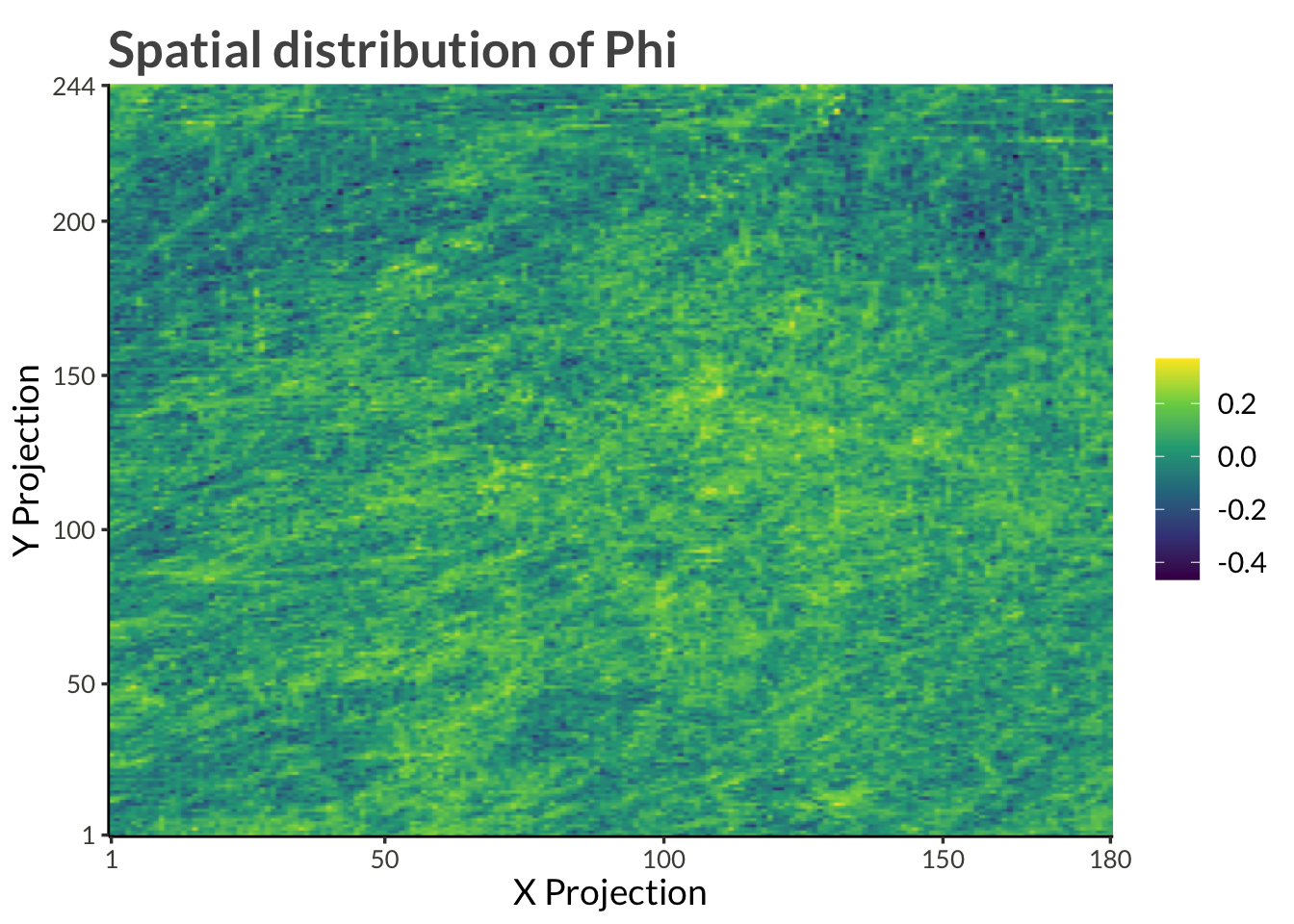
title = "Spatial distribution of Phi"
)
d |>
pivot_wider() |>
mutate(xi = link_shape_inverse(phi)) |>
ggplot(aes(proj_x, proj_y, fill = xi)) +
geom_raster(interpolate = TRUE) +
scale_x_continuous(
expand = expansion(),
breaks = c(range(d$proj_x), pretty(d$proj_x))
) +
scale_y_continuous(
expand = expansion(),
breaks = c(range(d$proj_y), pretty(d$proj_y))
) +
scale_fill_viridis_c() +
labs(
x = "X Projection",
y = "Y Projection",
fill = NULL,
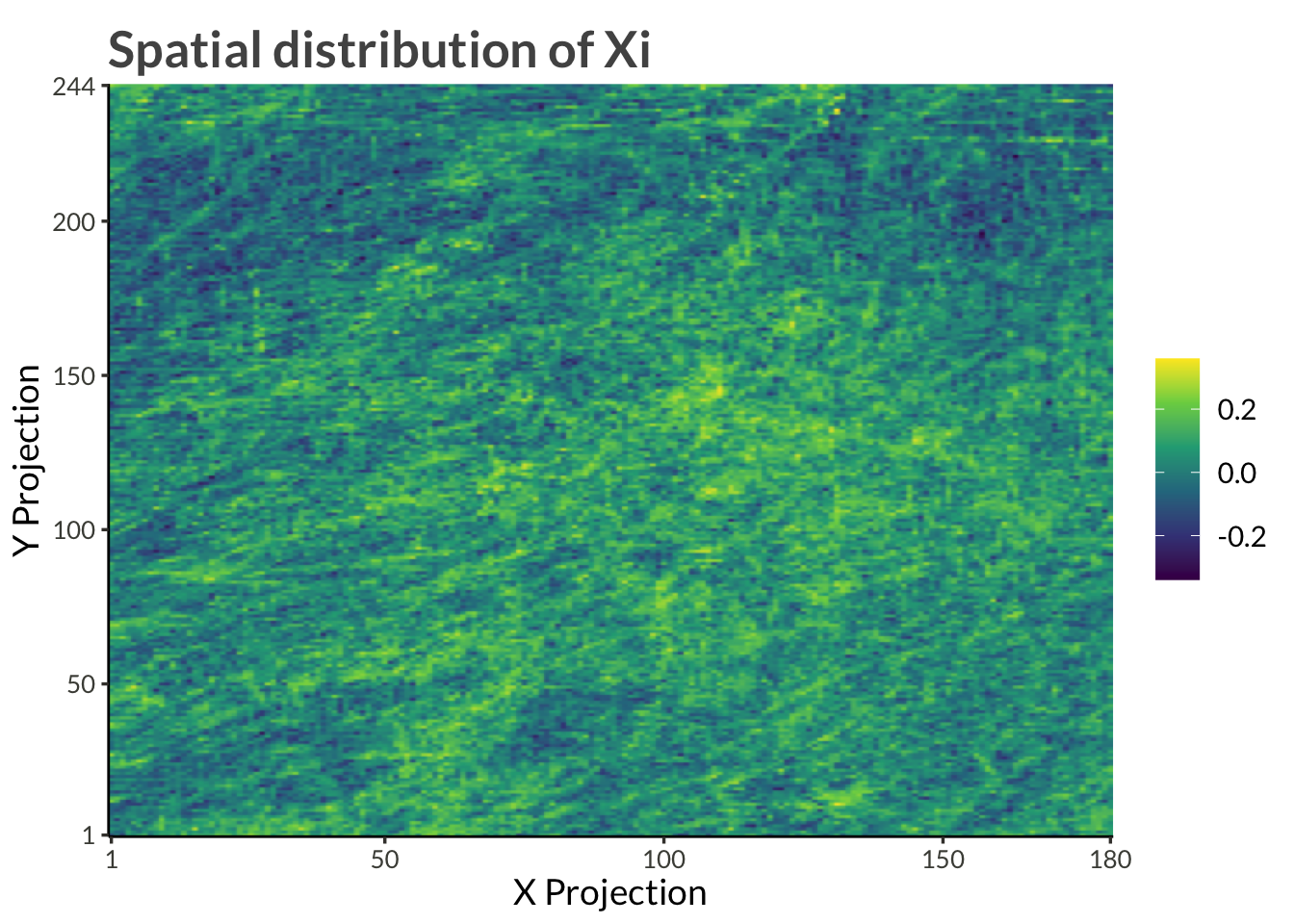
title = "Spatial distribution of Xi"
)
d |>
filter(name == "gamma") |>
ggplot(aes(proj_x, proj_y, fill = value)) +
geom_raster(interpolate = TRUE) +
scale_x_continuous(
expand = expansion(),
breaks = c(range(d$proj_x), pretty(d$proj_x))
) +
scale_y_continuous(
expand = expansion(),
breaks = c(range(d$proj_y), pretty(d$proj_y))
) +
scale_fill_viridis_c() +
labs(
x = "X Projection",
y = "Y Projection",
fill = NULL,
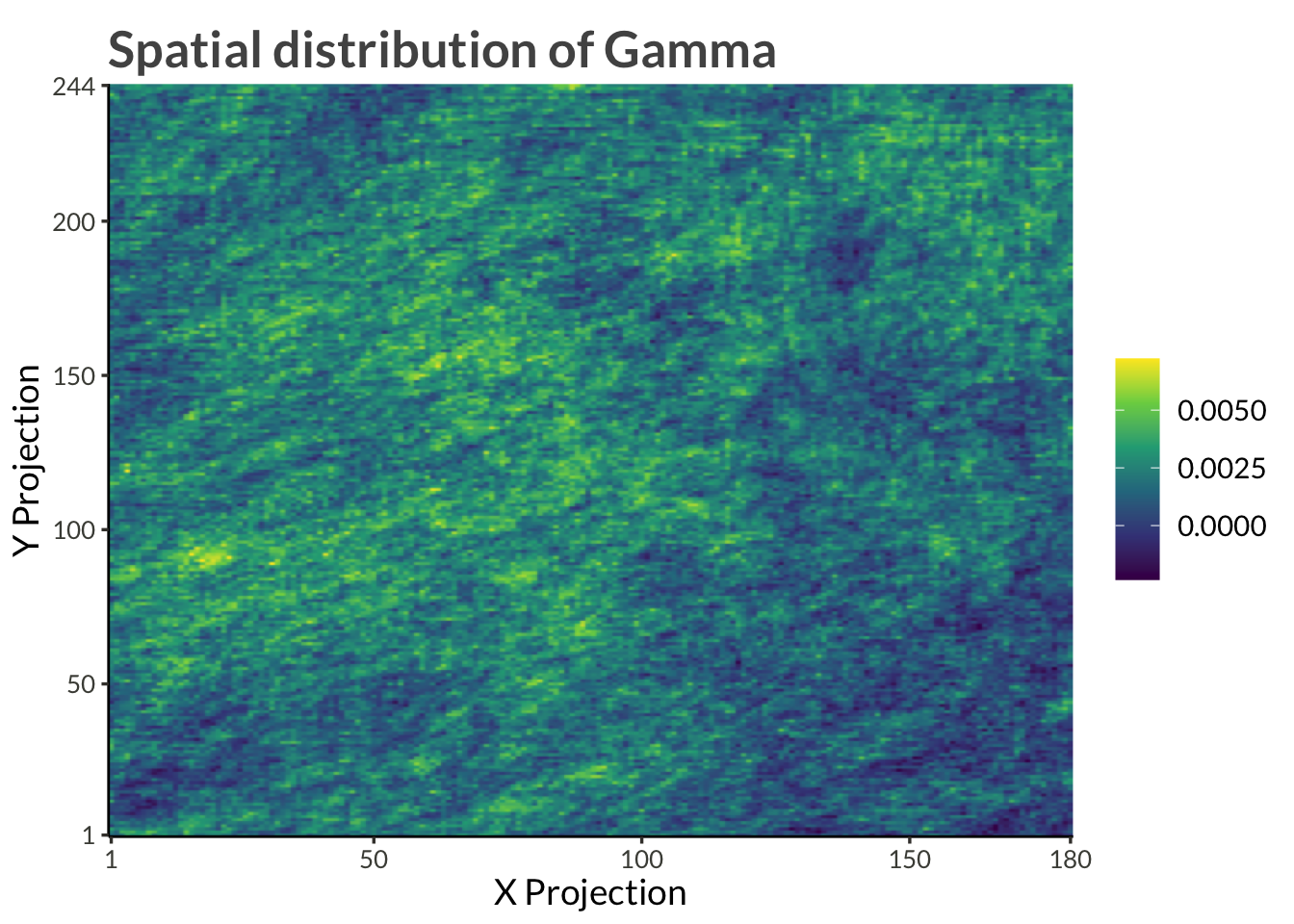
title = "Spatial distribution of Gamma"
)
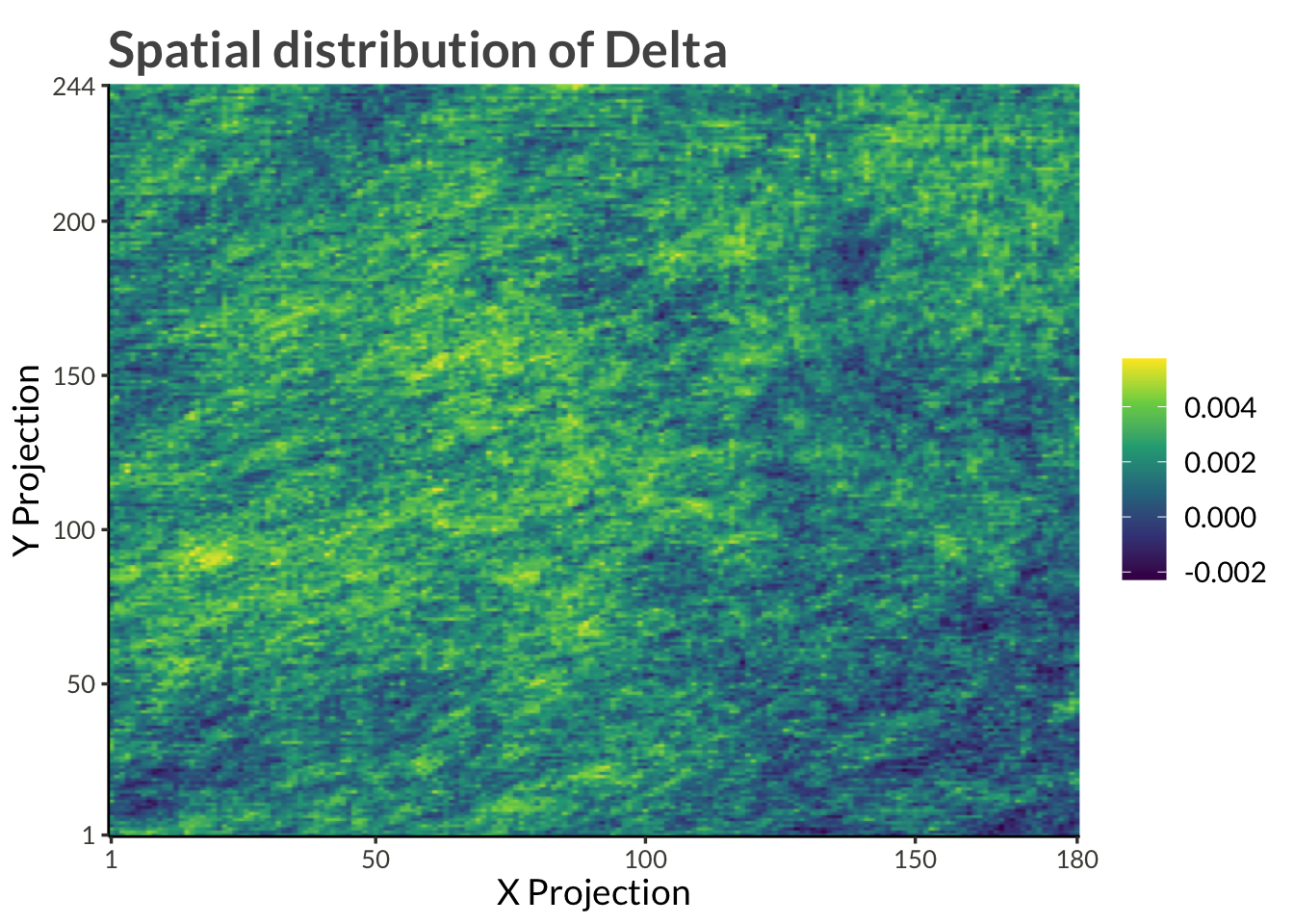
d |>
pivot_wider() |>
mutate(delta = link_trend_inverse(gamma)) |>
ggplot(aes(proj_x, proj_y, fill = delta)) +
geom_raster(interpolate = TRUE) +
scale_x_continuous(
expand = expansion(),
breaks = c(range(d$proj_x), pretty(d$proj_x))
) +
scale_y_continuous(
expand = expansion(),
breaks = c(range(d$proj_y), pretty(d$proj_y))
) +
scale_fill_viridis_c() +
labs(
x = "X Projection",
y = "Y Projection",
fill = NULL,
title = "Spatial distribution of Delta"
)
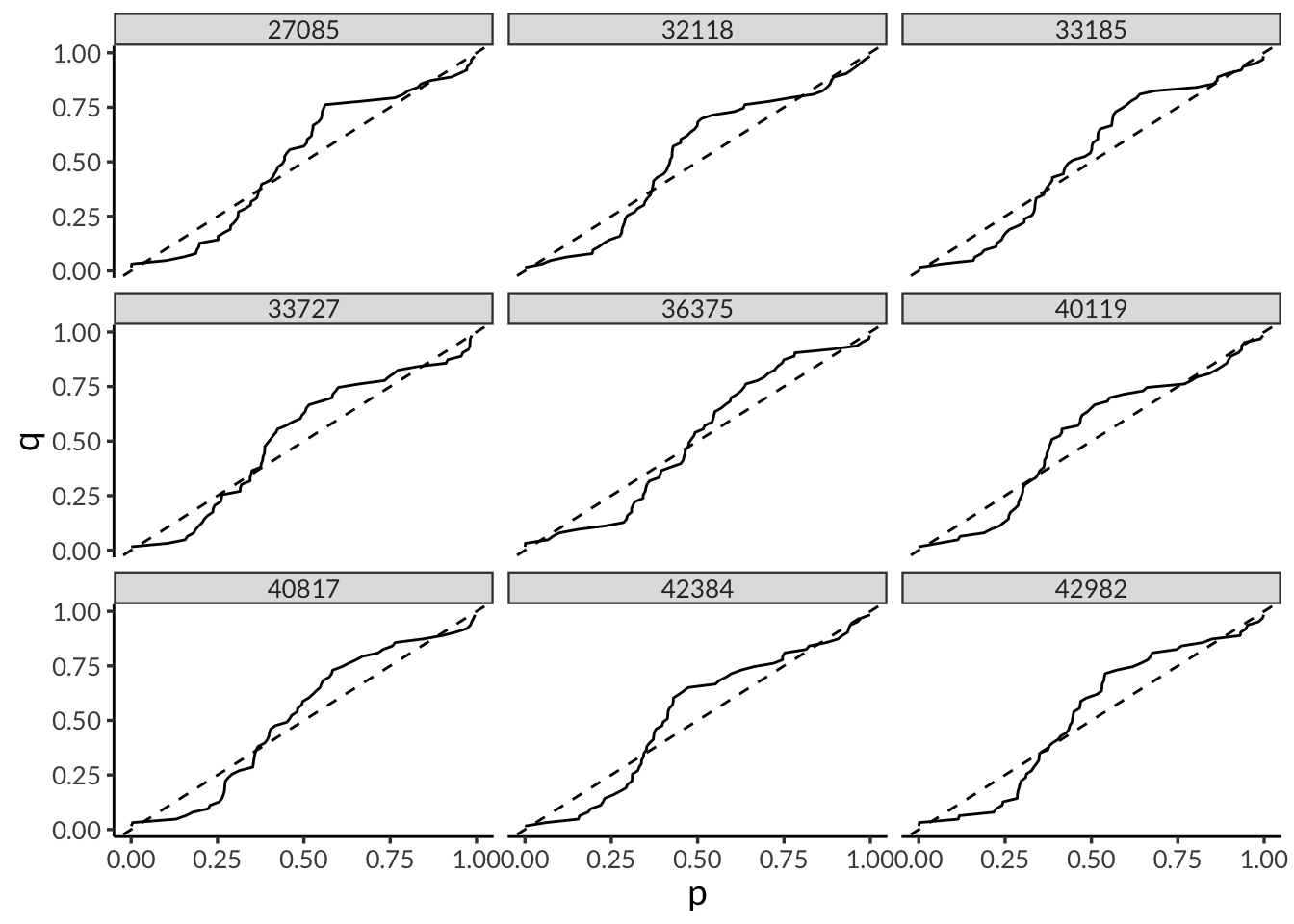
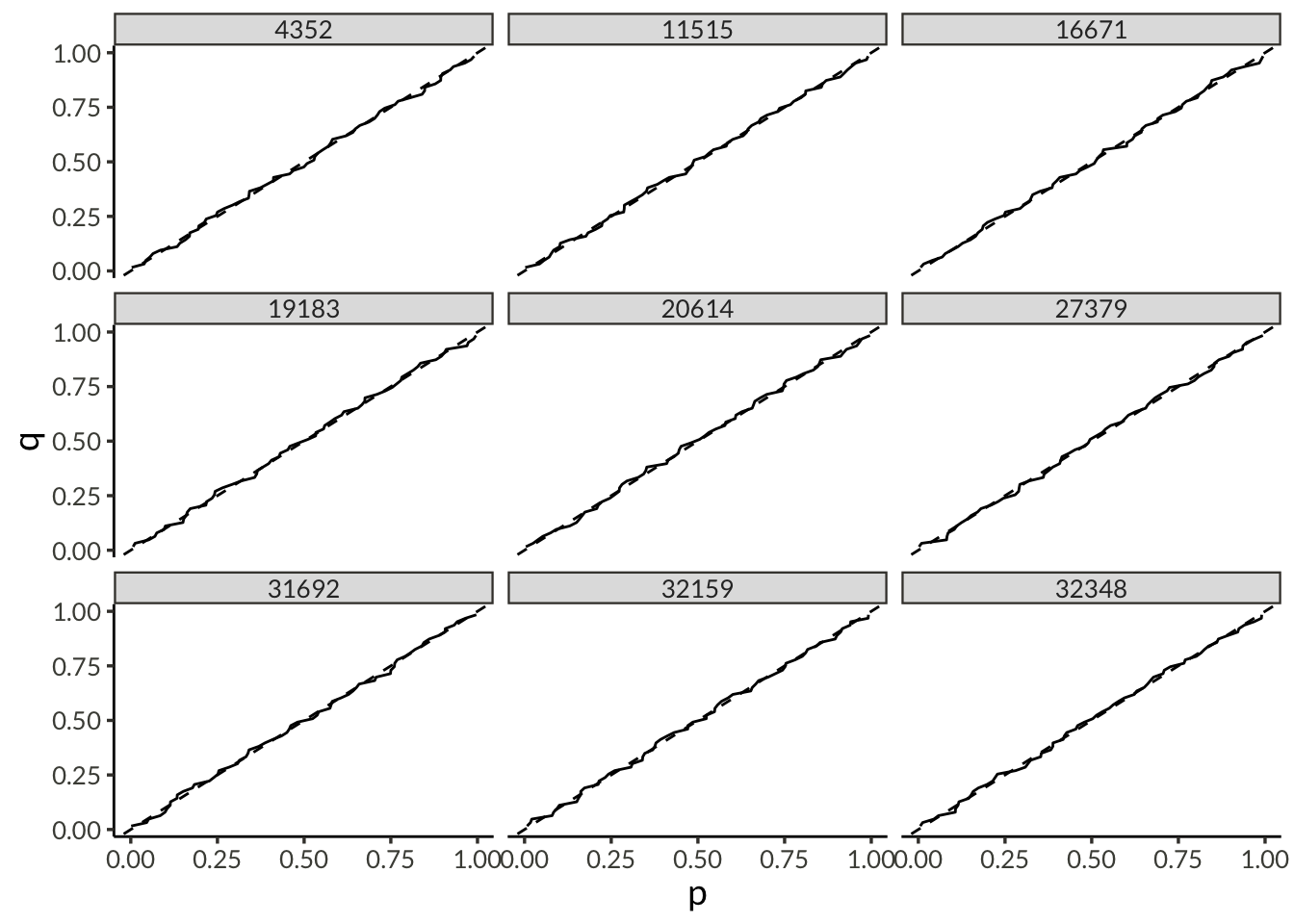
plot_dat <- d |>
pivot_wider(names_from = name, values_from = value) |>
mutate(
mu0 = exp(psi),
sigma = exp(tau + psi),
xi = link_shape_inverse(phi),
delta = link_trend_inverse(gamma)
) |>
select(-(psi:gamma)) |>
inner_join(
precip,
by = join_by(station),
multiple = "all"
) |>
mutate(
mu = mu0 * (1 + delta * (year - 1981)),
p = pgev(precip, mu, sigma, xi)
) |>
arrange(station, (precip - mu)) |>
select(station, year, precip, p) |>
mutate(
q = row_number() / (n() + 1),
station_diff = sqrt(mean((p - q)^2)),
.by = station
) |>
mutate(
year_diff = sqrt(mean((p - q)^2)),
.by = year
)